|
|
Provided here is a tool for sequence annotation directed PCR primer design targeting C. elegans promoter regions for the construction of promoter::GFP constructs using fusion PCR (PCR stitching). The design process is automated--only the gene name is required as input. Basic and advanced design parameters may be adjusted, and an optional e-PCR test is available to check primer specificity against the whole C. elegans genome.
|
|
|
|
|
|
|
|
|
| Basic Design Parameters |
- Gene : Enter the gene name for the promoter of interest, as found in Wormbase (under "Gene Name,Cosmid"). If the three-letter locus name is only known, check WormBase for the cosmid gene name.
|
- Number or primer sets : Number of external primer sets to be designed. Primers are listed by descending Primer Quality (Q value).
|
- Primer size range : Minimum size must be greater than zero and maximum size must be less than 35.
|
- Primer optimum Tm range : Melting temperature (Celcius) for a primer.
|
- PCR product size range : Primers will be designed to amplify a product (external, first round PCR) near the specified size range. Maximum size is 5 kb. By default, product size is reduced to avoid amplifying part of an upstream gene on the same strand. See Advanced Design Parameters below.
|
|
|
| Advanced Design Parameters (Optional) |
- GC clamp : Required number of G or C bases at the 3' of the left and right external primers. Default is 0.
|
- Max. Q Value : This is a PRIMER3 penalty score that indicates deviation from the specified optimal design parameters. Lower penalty score indicates a better primer pair. Deviations from the optimum primer size and Tm have a large influence on the penalty score. To read more about PRIMER3 and penalty score, follow the link below.
|
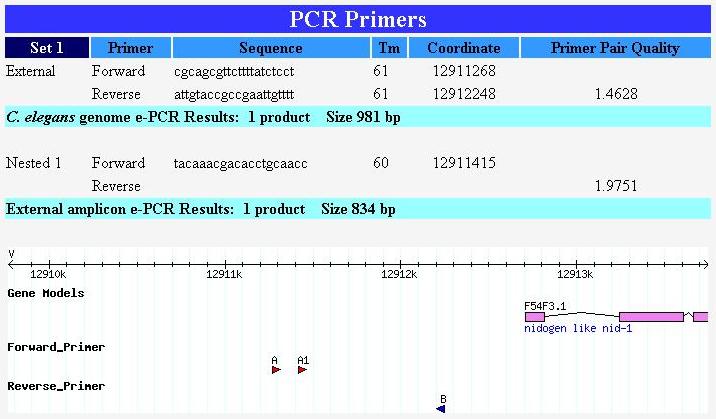
- Target upstream : Specify a target region rather than a product size. For the target start and target end, enter the number of base pairs upstream from the ATG start. In the example below, the target region entered is "500" to "1500." This will design primers to amplify a 1 kb region 500 bp upstream from the ATG start. Nested primers can also be designed for targeted regions.
Below: Partial output from a primer design request targeting a 1 kb region 500 bp upstream of the ATG start of gene F54F3.1.
|
- Allow primers in upstream neighbors : PCR product size may be reduced if a genes is found upstream, coding on the same strand (see Example output). Allowing primers in upstream neighbors forces a PCR product of the specified size, regardless of the presence of an upstream gene coding on the same strand.
|
- Exclude tandem repeats : Regions containing annotated tandem repeats can be excluded as potential primer sites. This option can be unchecked if Primer3 is having trouble picking primers; however, e-PCR is strongly recommended to check for multiple PCR products.
|
|
|
| Options for Nested PCR Primer Design |
|
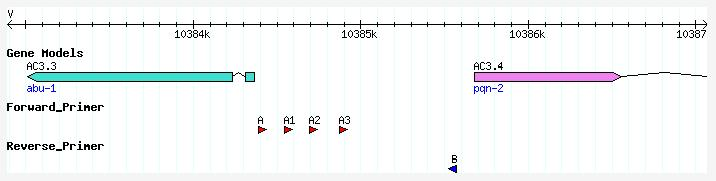
- Example : The image below shows the location of 2 external primers (A and B), designed to amplify the putative promoter region of gene AC3.4 in the first round of PCR. A series of 3 nested primers (A1, A2, and A3) were also designed. Using a different nested primer for the second round PCR will generate a series of promoter::GFP fusion constructs with progressively smaller regions of the original external amplicon.
|
|
- Primer separation : Specifiy the number of bases between the external primer and nested primer, or between sets of nested primers. In the example above all forward A primers are separated by approximately 150 bp.
|
|
|
|
| Primer Validation Using Electronic PCR |
- Default is 'ON'; this is slower but this test is recommended to avoid unexpected PCR prodcuts. External primers are checked against the whole C. elegans genome and nested primers are checked against the amplicon produced by the external primers. By default, e-PCR will search for products within 2000 bp of the expected product size. The stringency of the e-PCR check can be adjusted by changing the word size and allowed mismatches.
|
- Word size : Number of exact matches (W) at the 3' end of the primer. Default is 5. If a primer overlaps the ATG start of a gene, the primer is designed to mutate the ATG to ATC. The primer is designed so that the G->C mutation does not occur within 5 bp. of the 3' end of the primer.
|
- Allowed Mismatches : Maximum number of mismatches between the primer and the template, but not within W bases from the 3' end. Default is 2.
|
|
|
|
|
|
Sample Output |
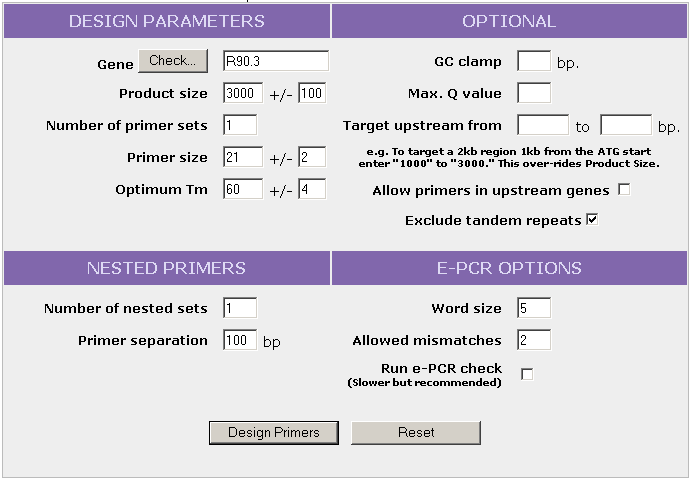
- Output from PCR fusion primer design : Gene information (location, coordinates, size, orientation), the location of the nearest upstream neighbor, and any adjustment to the requested external amplicon size (eg. to avoid an upstream neighbor) are indicated in the gene information section. A map shows the gene of interest in the context of surrounding genes. The primer design results table shows the primer sequences and coordinates, primer pair quality, primer Tm, and the expected PCR product size. If the e-PCR check is requested, the PCR product size(s) displayed will be the results from the e-PCR check. A primer map shows the location of the primers.
|
- Click on the image below to see the output generated from the request below.
|
 |
|
|
| Further Information |
- Primer3 documentation: WormPrimer uses Primer3 software (Rozen and Skaletsky, 1996, 1997, 1998) to design and pick primers. For a detailed description of Primer3 click here.
|
|
Last updated April 5, 2004
|